Scientific programming and performance of programming languages- A benchmarking example.
In this article, we will try to answer perhaps the most frequently asked question "Which programming language should I use for scientific programming?"
PROGRAMMING
Rahul Mahajan
9/1/20255 min read
Welcome back to this blog of scientific programming. In today’s article, I am going to throw a light on perhaps most frequently asked question i.e. “Which programming language should I use for scientific programming?” This is a question that I frequently came across during the course of interaction with my students at various occasions. And to be very honest, my answers to my students were strictly according to the problem requirements at the time.
Therefore, in this blog I will restrict myself from going into unnecessary details simply for two reasons. The first one is already given above and the second one is that, for most of the problems that I had come across in scientific programming, it is found that, if a problem is well defined and methodology is well understood, fifty percent problem is solved. For remaining fifty percent challenging part, we need to systematically frame set of computer instructions to which we call programming. You will be surprised to know that, statically typed compiled languages like C and Fortran can serve purpose of solving extremely complex problems and are being employed for this purpose even today. This can be understood from the fact that, legacy codes and procedures in scientific computations are still implemented in Fortran and C. Core libraries like LAPACK and BLAS are created using C and Fortran.
It doesn’t’ mean that the dynamically typed interpreted languages like Python, Julia, MATLAB are of less use. These languages offer superb functionalities and features which saves effort of writing many lines of code. In addition to this, they have a mature and vast ecosystem which offers many useful pre-built libraries ready to use for programmers.
In short, it’s a battle between performance vs functionalities. But is it always a scenario? Let’s do some exercise. I want you to install three languages in your system. C, Julia and Python.
Now open the IDE for each one of the languages and run the following codes snippets in appropriate IDEs.
/* Code for C language */
#include<stdio.h>
int main (void)
{
int i;
for(i=1;i<=1000;i++)
printf("%d\n",i);
return(0);
}
# code for Julia
for i in 1:1000
println(i)
end
# code for Python
for i in range(1,1001):
print (i)
These codes of few lines will make your computer to print numbers from 1 to 1000. Just note down the time taken by each of the languages to print the numbers on console. I insist to run these programs for at least two times without closing IDE and consider the average. (As sometimes, Julia may be sluggish in its first cold run)
While writing this post, I am using a very low-end processor. AMD Ryzen 3 3200U with Radeon Vega Mobile Gfx (2.60 GHz) and 8.0 GB RAM. The time required by each language to print 1000 number on standard output (i.e. console), in descending order is as follows.
Python>Julia>C.
And I hope that you have obtained same results. At this moment of time, if I ask, what language will you prefer from the perspective of execution speed? Certainly, at this moment of time, it is tempting to select C or Julia over Python. For computer, it was a very light weight task.
Let’s give a reasonably heavy task to computer.
In following three programs, I will be doing the following exercise.
First two 300 × 300 matrices are created with random numbers varying between 1 - 100. These matrices are multiplied for 300 times in a loop. In each loop iteration, the last element on major diagonal i.e element with index [300][300] is added to a variable ‘sum.’ Obviously, this variable ‘sum’ stores the addition of last diagonal elements of resultant matrix obtained from multiplication. Finally, the iterations count is printed using formula
N= sum/ K [300][300].
Where K [300][300] is the last element on major diagonal.
Below are the codes for C, Julia and python. Those who hate writing lengthy codes will prefer Julia or Python. But the previous example may force them to consider writing lengthy C code for the sake of speed. (Remember performance vs functionality)
You can download these codes from here.
Since we are not discussing the programming in detail here, at this moment of time, there is no need to delve into details of these codes like how they are written and why written so?
/* C Code */
#include<stdio.h>
#include<stdlib.h>
#include<time.h>
#define N 300
int main (void) {
int A[N][N], B[N][N], C[N][N];
int i, j, k, count = 1, iterations = 300;
long int sum = 0;
clock_t start, end;
double cpu_time;
for (i = 0; i < N; i++)
for (j = 0; j < N; j++) {
A[i][j] = (rand() % 100) + 1;
B[i][j] = (rand() % 100) + 1;
}
start = clock();
while (count <= iterations) {
for (i = 0; i < N; i++) {
for (j = 0; j < N; j++) {
C[i][j] = 0;
for (k = 0; k < N; k++) {
C[i][j] = C[i][j] + (A[i][k] * B[k][j]);
}
}
}
sum = sum + C[N - 1][N - 1];
count++;
}
end = clock();
cpu_time = ((double)(end - start) / CLOCKS_PER_SEC);
printf("CPU time: %.6f seconds\n", cpu_time);
printf("iterations are %f", (float)sum / C[N - 1][N - 1]);
getchar();
return 0;
}
# Julia Code
A=rand(1:100,300,300)
B=rand(1:100,300,300)
C=ones(300,300)
sum=0
@time begin
for i in 1:300
global C=A*B
global sum= sum+C[300,300]
end
end
println(" The number of iterations are ", sum/C[300,300])
# Python Code
import numpy as np
import time
counter =1
sum=0
A=np.random.randint(1, 101, size=(300, 300))
B=np.random.randint(1, 101, size=(300, 300))
start = time.process_time()
while counter<=300:
C=A*B
sum=sum+C[299,299]
counter=counter+1
end = time.process_time()
print("CPU time: {:.3f} seconds".format(end - start))
print("Iterations are :", sum/C[299,299])
The output shows the CPU time. The time for which the CPU was busy doing calculations. This time does not include user I/O time. (For Julia, we subtract the compilation time and garbage collection time from total time to get the result of CPU time.) After running these programs for 5 times, the table given below shows the result for average CPU Time for three languages. (On your system, the values will differ depending upon hardware you are using.)
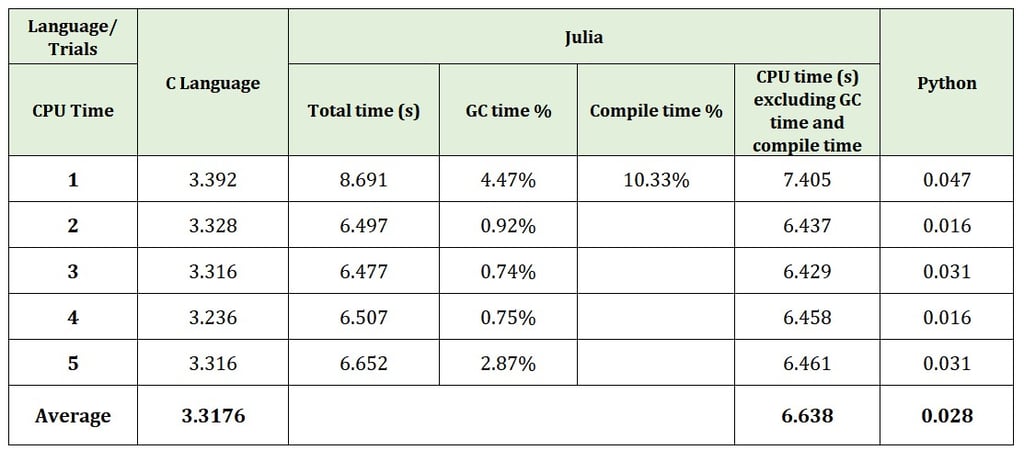
For getting speedy results, the compiler options for C were set as shown below.
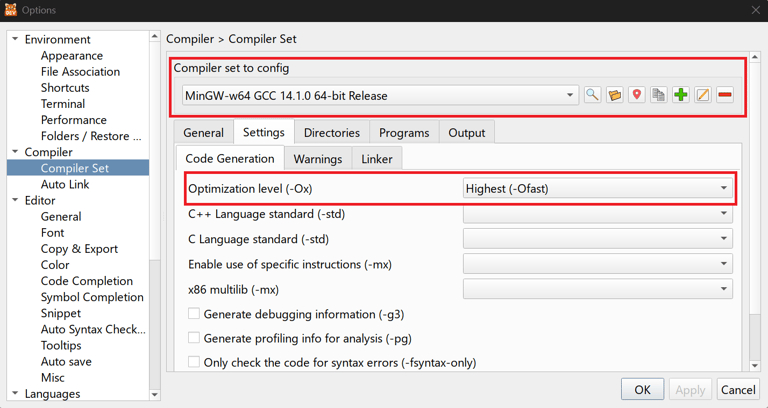
After looking at these results, certainly a question arises, what happened to python? What has given it a massive turbo boost?
Here, python has offered both performance (in terms of much less CPU Time) as well as functionality (by allowing us to write a concise code)
What happened here is the fact that, we haven’t simply used pure python. Instead, we used numpy library along with python. numpy is built with LAPACK and BLAS (written in C and Fortran) which are highly optimized for numerical computations. Thus, it was a competition between an unoptimized C algorithm vs highly optimized C Algorithm (not exactly but in a broader sense)
Final thought
If you prefer or rather love to have control over every minor detail in your program. I recommend you go with compiled languages like C / C++/ Fortran. If the problem is such that, it requires writing a lengthy code comprised of many lines, then use of dynamically typed interpreted languages like Python, Julia, Scilab can be a good choice. Alternatively, it is also possible to implement C/C++ for computationally intensive part of your program while for the remaining portion, dynamically typed languages like python can be implemented. There are various ways of doing this. We will further address such problems in our upcoming articles.
Leave a reply
scientificprogramming.in
Explore scientific programming
Contact author
© 2025. All rights reserved.
